What is autophagy?
The biological activities of eukaryotic cells are based on a delicate balance between synthesis and degradation. Since the discovery of DNA as the main body of genetic information, protein synthesis systems through transcription and translation have been widely studied. However, it is easy to imagine that synthesis alone would flood the cell with proteins and adversely affect the cell itself. In this context, research on proteolytic systems has been attracting attention in recent years. Along with the ubiquitin-proteasome system, which selectively degrades short-lived proteins, a system called autophagy (autophagy) contributes significantly to the degradation of long-lived proteins. Autophagy is a widely conserved system in eukaryotes that degrades cytoplasmic components on a large scale via vacuoles/lysosomes.
Molecular mechanism of autophagosome formation
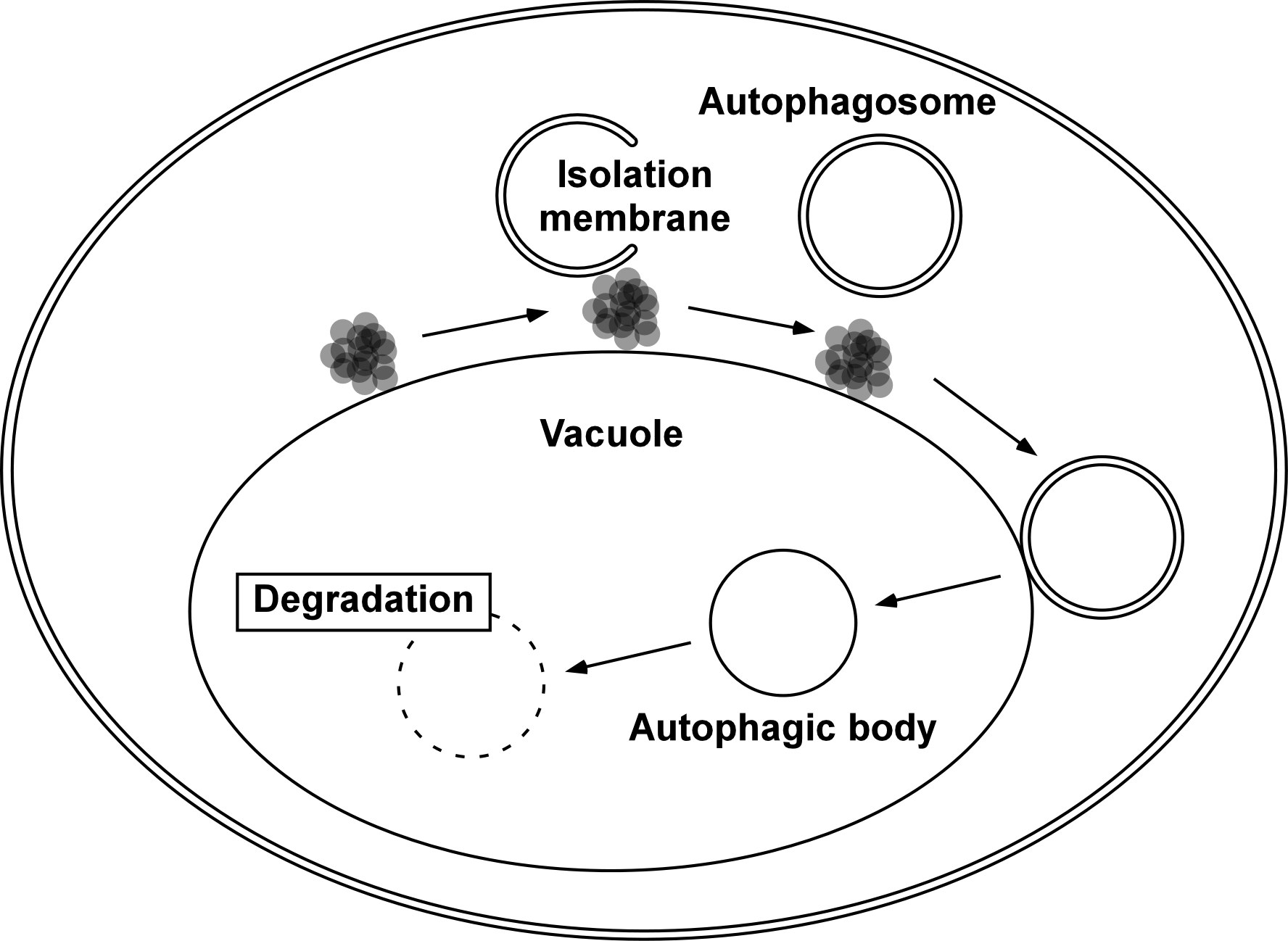
When autophagosome formation is induced by nutrient starvation, a cup-shaped membrane structure called an isolation membrane appears in the cytoplasm. The isolation membrane curves and extends to surround the components to be degraded, and eventually becomes a double membrane organelle, an autophagosome (Fig. 1). In our laboratory, autophagy has been studied using the budding yeast Saccharomyces cerevisiae as a model system for eukaryotic cells. Autophagy is an interesting phenomenon because the process of autophagosome formation proceeds with a very characteristic and dynamic membrane phenomenon. We are studying this issue using live imaging techniques with fluorescence microscopy.
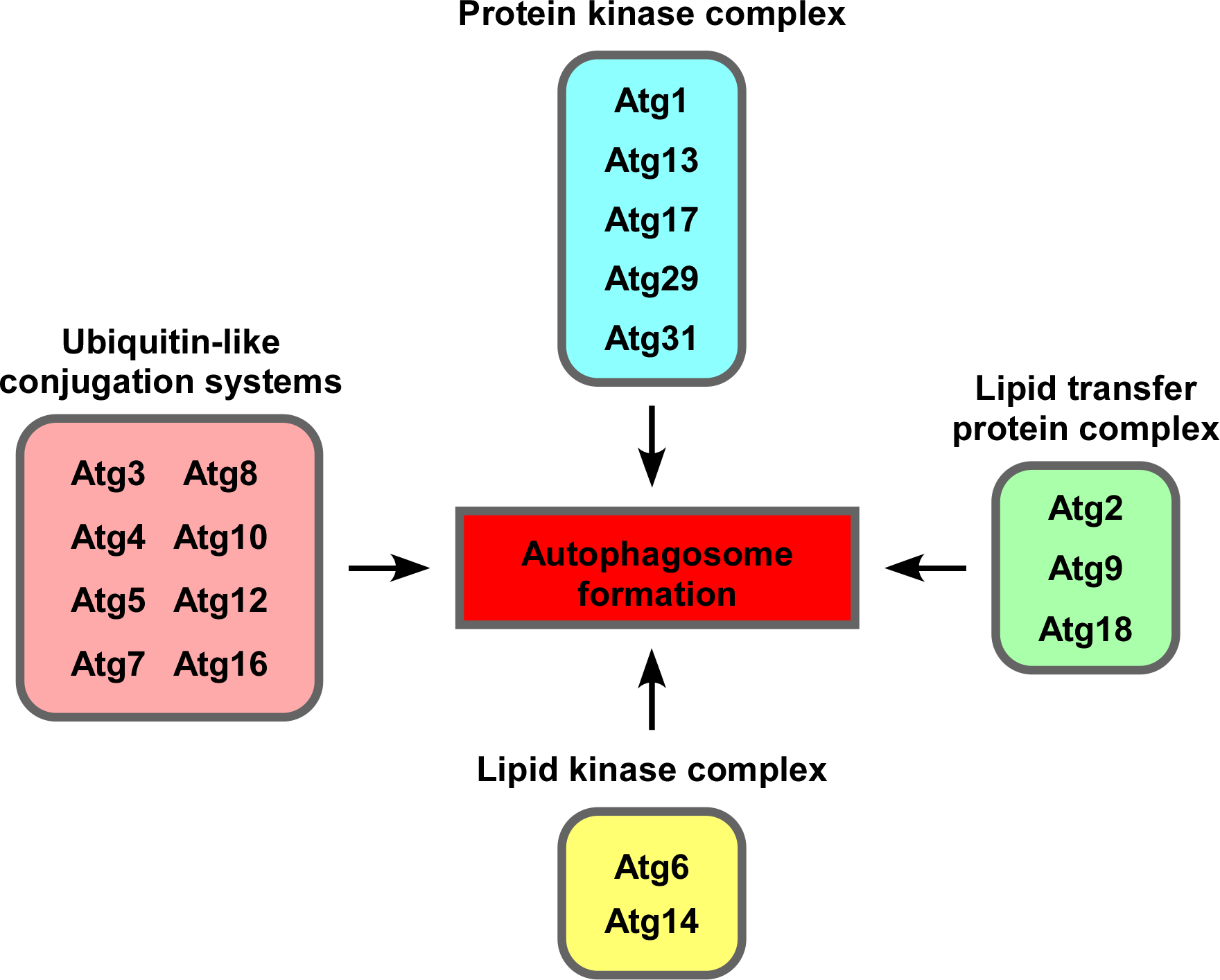
Eighteen autophagy-related (Atg) proteins are involved in autophagosome formation. They also function as a group of proteins (Fig. 2). Our laboratory has developed a method for visualization of the isolation membrane of budding yeast and has captured the subcellular localization of each Atg protein during the formation of the membrane by live imaging and quantitative analysis of the obtained image data to elucidate the entire molecular mechanism of autophagosome formation.
Molecular mechanism of autophagic body degradation
Autophagosomes formed in the cytoplasm fuse with the vacuole, an intracellular degradation compartment, and release an autophagic body composed of an inner membrane compartment into the vacuole. The autophagic body is degraded by intravacuolar hydrolytic enzymes. In budding yeast, the progression of autophagy can be monitored using an light microscope. Autophagic bodies can be seen as dozens of particles in the vacuole (Fig. 3). Degradation of autophagic bodies requires a lipase called Atg15, along with proteases called Pep4 and Prb1. We are studying the process of autophagic body degradation by Atg15 to reconstruct and visualize it by live imaging.
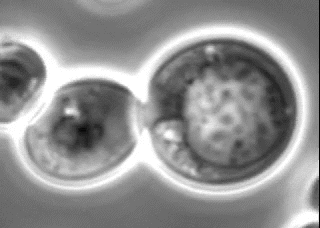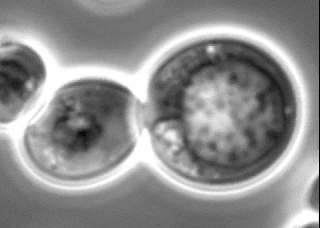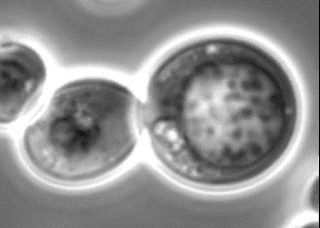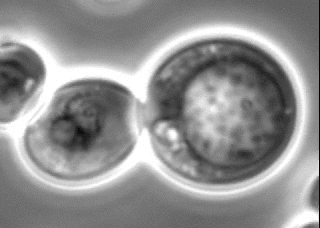
Search for novel membraneless organelles
The nucleolus has long been known as a membraneless organelle. In recent years, several membraneless organelles have been reported. However, a comprehensive screening has not yet been conducted. We have been conducting a genome-wide screening for novel membraneless organelles from an original perspective. We are now conducting a detailed analysis of the obtained candidates from a cell biological viewpoint.
